Nanopore Adaptive Sampling Revolutionizes Target Enrichment
Overview of Adaptive Sampling
In a groundbreaking advancement for genomics, the concept of Nanopore Adaptive Sampling has emerged as a transformative strategy, pushing the boundaries of target enrichment. This novel approach introduces a paradigm shift by seamlessly integrating target enrichment or depletion directly into the sequencing process. This eliminates the intricate target enrichment steps typically required during initial sample preparation. At the core of this advancement lies its integration into the pivotal MinKNOW operating software, which governs all nanopore sequencing devices.
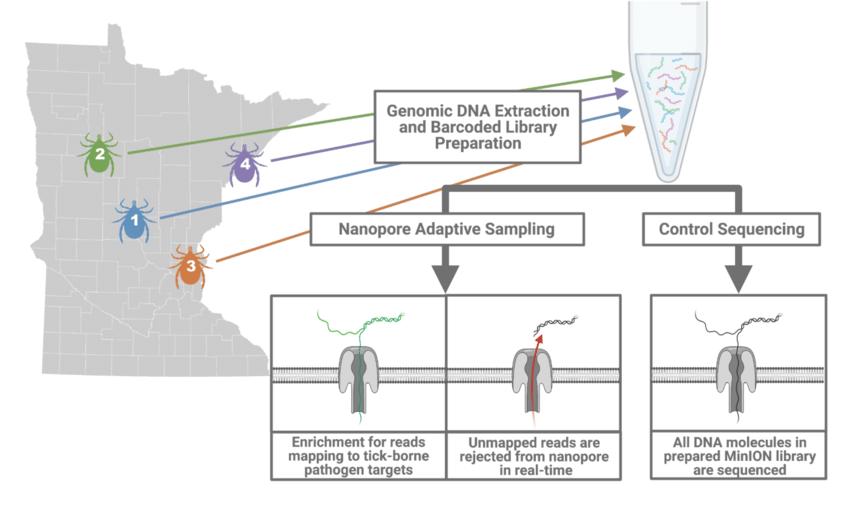
Comparison of nanopore adaptive sampling (NAS) and control sequencing methods. (Kipp et al., 2021)
The Adaptive Sampling Workflow
The operational workflow commences by meticulously preparing the entire DNA library for sequencing, scrupulously avoiding any amplification or enrichment steps. Once ready, the sample is introduced into the flow cell, and the sequencing process is meticulously configured with the software. Here, the game-changing element of adaptive sampling is activated, substantiated by the uploading of a reference file in FASTA format and a BED file housing the coordinates pertinent to enrichment or depletion.
Subsequently, the sequencing run is initiated as customary, marking the commencement of the sequencing journey. Real-time sequencing reveals its prowess by enabling the instantaneous identification of sequence relevance as the DNA strand traverses the nanopore. The promptly aligning the initial segment of the sequenced strand with the provided reference accomplished this impressive feat
The Software Algorithm
The ingenious software algorithm takes center stage in decision-making: if the examined sequence aligns with a designated enrichment target or does not correspond to a depletion sequence, the software allows the sequencing process to continue. Conversely, if the sequence does not align with an intended target or is marked for depletion, the software triggers the selective ejection of the strand from the nanopore. This intelligent maneuver effectively stops further sequencing of irrelevant sequences, optimizing pore occupancy time for areas of significant interest.
Notably, this transformative technique is not bound by limitations in read length or the number of targets. This exceptional flexibility empowers the comprehensive enrichment or depletion of extensive panels, or even entire genomes, during the sequencing process itself. As a result, the conventional reliance on labor-intensive and time-consuming lab-based procedures like hybridization capture becomes obsolete.
The advent of Nanopore Adaptive Sampling carries profound implications for the genomics field. By seamlessly integrating target enrichment or depletion into the sequencing process, researchers can expedite the identification of regions of interest. This not only accelerates research timelines but also holds the potential to revolutionize genomics methodologies, paving the way for novel discoveries and insights. As this transformative approach gains further traction and evolves, it is likely to become a cornerstone of modern genomics research, transcending existing limitations and unlocking new realms of scientific exploration.
In summary, the advent of Nanopore Adaptive Sampling stands as a testament to the fusion of bioinformatics innovation and real-time nanopore sequencing. Its seamless integration within the sequencing process, obviating the need for cumbersome target enrichment during sample preparation, marks a pivotal stride toward streamlined and efficient genomics research. This pioneering strategy not only expedites the identification of regions of interest but also holds the potential to revolutionize genomics research methodologies as we know them.
Learn other:
As a leading genomics company, CD Genomics provides next-generation sequencing and bioinformatics services to pharmaceutical and biotech companies, as well as academic and government agencies around the world, relying on advanced sequencing instruments and rich project experience. The next era of DNA sequencing technology, also known as termed long-read sequencing or third-generation sequencing (TGS), plays an increasingly important role in biology. We are offering long-read sequencing services from PacBio and Oxford Nanopore, giving researchers a wide range of cutting-edge sequencing services to suit particular needs at every stage of any projects. With years of experience in long-read sequencing technology, we are able to generate and analyze high quality data using the latest workflows.
Nanopore sequencing with Oxford Nanopore Technologies (ONT) systems is able to perform high-throughput long-read sequencing of both DNA and RNA samples, involving genomic DNA, amplified DNA, cDNA, and RNA. Briefly, the nanopore sequencer is based on a novel ‘nanopore technology that uses nanopores embedded in an electro-resistant membrane. A voltage is set on the membrane to form an ionic current across the pores. When a DNA molecule passes through a nanopore, the current variation, producing a characteristic signal, which is mainly influenced by four DNA bases. Then a base-calling algorithm converts the current variation back into the original DNA sequence in real-time.
Reference
- Kipp, Evan J., et al. “Enabling metagenomic surveillance for bacterial tick-borne pathogens using nanopore sequencing with adaptive sampling.” BioRxiv (2021): 2021-08.











