Antibody Heavy and Light Chains: Structure and Function
Antibodies, or immunoglobulins, are crucial components of the immune system, tasked with identifying and neutralizing foreign objects such as bacteria and viruses. These sophisticated molecules exhibit remarkable specificity and diversity, largely attributable to their intricate structure comprising heavy and light chains. This article delves into the structural and functional nuances of antibody heavy and light chains, elucidating their pivotal roles in immune response and therapeutic applications.
Basic Structure of Antibodies
Antibodies exhibit a characteristic Y-shaped structure composed of four polypeptide chains: two identical heavy (H) chains and two identical light (L) chains. This structural arrangement is essential for their diverse functions in immune defense.
Composition of Antibodies
Heavy Chains (H Chains):
- Structure: Each antibody molecule contains two heavy chains, which are longer and more complex than light chains. Heavy chains are encoded by gene segments known as V (variable), D (diversity), and J (joining), allowing for significant diversity in antibody structure and function.
- Domains: Heavy chains consist of several domains, including a variable domain (VH) at the N-terminus and one or more constant domains (CH1, CH2, CH3, and sometimes CH4) towards the C-terminus. The VH domain is crucial for antigen binding and recognition due to its variability, determined by the V, D, and J gene segments.
Light Chains (L Chains):
- Structure: Antibodies also contain two light chains, which are shorter and less complex than heavy chains. Light chains are encoded by gene segments V and J, similar to the V and J segments of heavy chains.
- Domains: Light chains possess a variable domain (VL) and a constant domain (CL). The VL domain contains three hypervariable regions, also known as complementarity-determining regions (CDRs), which directly interact with antigens.
Y-Shaped Structure
The combination of heavy and light chains gives rise to the characteristic Y-shaped structure of antibodies, which is crucial for their function. The Y shape consists of two identical antigen-binding fragments (Fab) at the tips of the Y and one constant fragment (Fc) at the base of the Y.
- Fab Fragments: Each Fab fragment consists of one variable domain from a heavy chain (VH) and one variable domain from a light chain (VL), forming the antigen-binding site. The specificity of antibody-antigen interactions is primarily determined by the unique combination of VH and VL domains.
- Fc Fragment: The Fc fragment is composed of constant domains from the heavy chains (CH2 and CH3). It determines the antibody’s class (e.g., IgG, IgA, IgM) and mediates interactions with other components of the immune system, such as complement proteins and Fc receptors on immune cells.
Disulfide Bonds and Stability
Antibodies are stabilized by disulfide bonds, covalent linkages formed between cysteine residues within and between the heavy and light chains. These bonds contribute to the structural integrity of antibodies, ensuring they remain functional under various physiological conditions.
Flexibility and Functionality
The hinge region between the Fab and Fc fragments provides flexibility to the antibody molecule, allowing each Fab fragment to move independently. This flexibility is essential for antibodies to bind to antigens with different spatial orientations and sizes, enhancing their efficacy in recognizing and neutralizing diverse pathogens.
Diagram of Antibody Structure
Below is a simplified diagram illustrating the basic structure of an antibody:
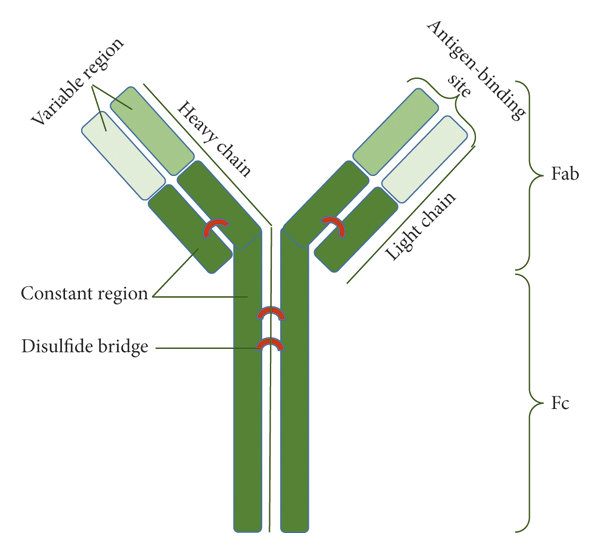
A schematic representation of the antibody structure (Golpour ET AL., 2021).
Structure of Heavy Chains
Gene Segments and Diversity of Heavy Chains
V (Variable), D (Diversity), and J (Joining) Segments: Heavy chains are encoded by genes that undergo somatic recombination of these segments during B cell development. This process generates extensive diversity in the variable regions of antibodies, enabling recognition of a wide range of antigens.
Types of Heavy Chains: Humans have five types of heavy chains, each associated with a specific class of antibody (Ig):
- α (IgA), δ (IgD), ε (IgE), γ (IgG), and μ (IgM): These classes differ in their constant region sequences, which dictate their biological functions, such as opsonization, neutralization, and complement activation.
Structural Domains of Heavy Chains
Variable Region (VH): Located at the N-terminal end of each heavy chain, the VH domain is highly variable due to differences in the V, D, and J gene segments. It contains three complementarity-determining regions (CDRs) that directly interact with antigens, determining the specificity of antibody binding.
Constant Regions (CH1, CH2, CH3, and sometimes CH4): Found towards the C-terminal end of heavy chains, these constant domains exhibit less variability across antibody classes but are crucial for effector functions and antibody stability.
Structural Features and Flexibility
Hinge Region:
Located between the Fab and Fc fragments of antibodies, the hinge region of heavy chains provides flexibility and mobility to the antigen-binding arms. This flexibility allows antibodies to engage antigens at various angles and orientations, enhancing their effectiveness in immune defense.
Glycosylation:
Heavy chains, like many glycoproteins, undergo glycosylation, where carbohydrate molecules are attached to specific amino acid residues. These glycans can modulate antibody stability, solubility, and interactions with other immune components, influencing overall immune response dynamics.
Function and Importance of Heavy Chains
Antigen Binding:
The VH domain of heavy chains, along with the variable domain (VL) of light chains, forms the antigen-binding site of antibodies. This region contains three hypervariable loops, known as complementarity-determining regions (CDRs), which directly interact with antigens.
CDRs in the VH domain confer specificity to the antibody by recognizing and binding to unique epitopes on antigens, such as proteins on pathogens or abnormal cells.
Heavy chains contribute significantly to the affinity and specificity of antibody-antigen interactions. Through somatic recombination of V, D, and J gene segments, heavy chains generate a diverse repertoire of VH domains that can recognize a broad range of antigens with varying affinities.
Effector Functions:
Class Switching: Heavy chains determine the class or isotype of antibodies (e.g., IgG, IgA, IgM) through a process called class switching. This process occurs after initial antigen exposure and is influenced by cytokines and other signals from immune cells. Different antibody classes have distinct effector functions mediated by their constant regions, such as opsonization (IgG), mucosal immunity (IgA), and allergic responses (IgE).
Effector Mechanisms: The constant regions (CH2 and CH3) of heavy chains interact with complement proteins and Fc receptors on immune cells, initiating effector mechanisms that eliminate pathogens or infected cells. For example, IgG antibodies can activate the complement cascade, leading to pathogen lysis, or engage Fc receptors on phagocytes to facilitate pathogen clearance through phagocytosis.
Structural Stability:
The constant domains of heavy chains, particularly CH2 and CH3, contribute to the overall stability and structural integrity of antibodies. Disulfide bonds between cysteine residues within these domains reinforce the antibody’s tertiary structure, ensuring functionality in diverse physiological environments.
Detailed Structure of Light Chains
Gene Segments and Diversity of Light Chains
V (Variable) and J (Joining) Segments: Light chains are encoded by genes that undergo somatic recombination of these segments during B cell development. This process generates diversity in the variable regions of antibodies, enabling them to recognize a wide range of antigens.
Types of Light Chains: There are two main types of light chains found in antibodies:
- Kappa (κ) Chains: The majority of antibodies (around 60-70%) in humans have kappa light chains.
- Lambda (λ) Chains: The remaining antibodies (around 30-40%) have lambda light chains.
Both kappa and lambda light chains perform similar functions in antigen recognition and binding, but their genetic origins and distributions differ.
Structural Domains of Light Chains
Variable Region (VL): Located at the N-terminal end of each light chain, the VL domain contains three hypervariable regions, also known as complementarity-determining regions (CDRs). These CDRs directly interact with antigens, determining the specificity and affinity of antibody binding.
Constant Region (CL): The CL domain is found towards the C-terminal end of light chains and provides structural stability to the light chain.
Comparative Analysis of Kappa and Lambda Chains
Distribution and Functionality:
- Kappa (κ) chains are more prevalent in antibodies produced by B cells, accounting for approximately 60-70% of total antibodies in humans.
- Lambda (λ) chains are less common, comprising the remaining 30-40% of antibodies.
- Despite this difference in prevalence, both types of light chains contribute equally to antibody function and diversity in antigen recognition.
Genetic Diversity:
The genetic mechanisms that govern the expression of kappa and lambda light chains provide additional diversity in antibody repertoire, enabling the immune system to respond effectively to a wide array of pathogens and antigens.
Function and Importance of Light Chains
Antigen Binding:
The VL domain of light chains contains three hypervariable loops, or complementarity-determining regions (CDRs), which directly interact with antigens. CDRs in the VL domain, in conjunction with CDRs in the VH domain of heavy chains, determine the specificity and affinity of antibody-antigen interactions. Through somatic recombination of V and J gene segments, light chains generate a diverse repertoire of VL domains that can recognize a broad range of antigens with high specificity.
Stability and Flexibility:
Light chains contribute to the overall stability and flexibility of the antibody molecule. The interaction between light and heavy chains allows antibodies to adopt different conformations, optimizing antigen recognition and binding.
Pairing with Heavy Chains:
Each light chain interacts with one heavy chain to form a functional antigen-binding site. This pairing involves complementary interactions between the variable regions of both chains, ensuring precise and effective antigen recognition.
Analysis Methods for Antibody Heavy Chains and Light Chains
Antibody Sequencing
- Mass Spectrometry (MS) Antibody Sequencing: Utilizes high-resolution mass spectrometry to sequence peptides derived from enzymatic digestion of antibody heavy and light chains. This method provides detailed information about the amino acid sequence, post-translational modifications, and structural variants.
- Edman Degradation: A classical method for sequencing proteins, involving stepwise cleavage of amino acids from the N-terminus of the chain. Though less commonly used now due to automation and sensitivity limitations, it still provides reliable sequence data.
Protein Structure Analysis
- X-ray Crystallography: Determines the three-dimensional structure of antibody heavy and light chains by analyzing diffraction patterns of X-rays passing through crystallized protein samples. Provides atomic-level details of antibody-antigen interactions and conformational changes.
- Cryo-Electron Microscopy (Cryo-EM): Uses electron microscopy to visualize frozen hydrated specimens of antibody complexes. Allows for the determination of antibody structure at near-atomic resolution without the need for crystallization.
Functional Assays
- Enzyme-Linked Immunosorbent Assay (ELISA): Quantifies and detects specific antibody heavy and light chains based on their interaction with antigens immobilized on a solid surface. Used for assessing antibody binding affinity, specificity, and concentration.
- Surface Plasmon Resonance (SPR): Measures real-time interactions between antibody heavy and light chains and antigens immobilized on a sensor surface. Provides kinetic data on binding affinity, association, and dissociation rates.
Bioinformatics and Computational Modeling
- Homology Modeling: Predicts the three-dimensional structure of antibody heavy and light chains based on known structures of related proteins. Useful for understanding structure-function relationships and designing engineered antibodies.
- Sequence Alignment and Phylogenetic Analysis: Compares antibody heavy and light chain sequences across species or within individuals to identify conserved regions, mutations, and evolutionary relationships. Helps elucidate antibody diversity and evolution.
Reference
- Golpour, Monireh, et al. “The Perspective of Therapeutic Antibody Marketing in Iran: Trend and Estimation by 2025.” Advances in pharmacological and pharmaceutical sciences 2021.1 (2021): 5569590.
Read More: Role of Phosphatidic Acid in Lipid Metabolism








